 |
|
|
[Sponsors] | |||||
How to calculate the Chemical Time Scale using Jacobian Matrix Eigenvalues? |
 |
|
|
LinkBack | Thread Tools | Search this Thread | Display Modes |
|
|
|
#1 |
|
New Member
Harsh Anand
Join Date: May 2024
Posts: 12
Rep Power: 2  |
Hi Everyone!!
How do I calculate the chemical time scale in the PaSR Model using the theory of eigenvalues of the Jacobian Matrix? A/c to Fox (https://books.google.co.in/books?hl=...page&q&f=false) and https://doi.org/10.1016/j.apenergy.2018.04.085, the chemical time scale can be computed by following the below process: 1. Compute the Jacobian matrix: 2. After the decomposition of the Jacobian matrix, the chemical time scale is estimated with the inverse of the eigenvalues 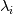 as: as:where, 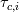 is the characteristic time scale of a single species. is the characteristic time scale of a single species.3. After removing the dormant species (characterised by infinite time scale values), the slowest chemical time scale is chosen as the leading scale for the evaluation of the PaSR parameter  . .Now, how do I calculate this Jacobian Matrix and its eigenvalues in the PaSR.C File? I am aware that a Jacobian Member Function is available in the StandardChemistryModel.C File, but since I am new to C++ and programming, I don't know how to compute the Jacobian and subsequently, its eigenvalues. Any help is appreciated. PaSR.C File: Code:
/*---------------------------------------------------------------------------*\
========= |
\\ / F ield | OpenFOAM: The Open Source CFD Toolbox
\\ / O peration |
\\ / A nd | Copyright (C) 2011-2017 OpenFOAM Foundation
\\/ M anipulation |
-------------------------------------------------------------------------------
License
This file is part of OpenFOAM.
OpenFOAM is free software: you can redistribute it and/or modify it
under the terms of the GNU General Public License as published by
the Free Software Foundation, either version 3 of the License, or
(at your option) any later version.
OpenFOAM is distributed in the hope that it will be useful, but WITHOUT
ANY WARRANTY; without even the implied warranty of MERCHANTABILITY or
FITNESS FOR A PARTICULAR PURPOSE. See the GNU General Public License
for more details.
You should have received a copy of the GNU General Public License
along with OpenFOAM. If not, see <http://www.gnu.org/licenses/>.
\*---------------------------------------------------------------------------*/
#include "PaSR.H"
// * * * * * * * * * * * * * * * * Constructors * * * * * * * * * * * * * * //
template<class ReactionThermo>
Foam::combustionModels::PaSR<ReactionThermo>::PaSR
(
const word& modelType,
ReactionThermo& thermo,
const compressibleTurbulenceModel& turb,
const word& combustionProperties
)
:
laminar<ReactionThermo>(modelType, thermo, turb, combustionProperties),
Cmix_(this->coeffs().getScalar("Cmix")),
kappa_
(
IOobject
(
thermo.phasePropertyName(typeName + ":kappa"),
this->mesh().time().timeName(),
this->mesh(),
IOobject::NO_READ,
IOobject::AUTO_WRITE
),
this->mesh(),
dimensionedScalar(dimless, Zero)
)
{}
// * * * * * * * * * * * * * * * * Destructor * * * * * * * * * * * * * * * //
template<class ReactionThermo>
Foam::combustionModels::PaSR<ReactionThermo>::~PaSR()
{}
// * * * * * * * * * * * * * * Member Functions * * * * * * * * * * * * * * //
template<class ReactionThermo>
void Foam::combustionModels::PaSR<ReactionThermo>::correct()
{
if (this->active())
{
laminar<ReactionThermo>::correct();
tmp<volScalarField> tepsilon(this->turbulence().epsilon());
const scalarField& epsilon = tepsilon();
tmp<volScalarField> tmuEff(this->turbulence().muEff());
const scalarField& muEff = tmuEff();
tmp<volScalarField> ttc(this->tc());
const scalarField& tc = ttc();
tmp<volScalarField> trho(this->rho());
const scalarField& rho = trho();
forAll(epsilon, i)
{
const scalar tk =
Cmix_*sqrt(max(muEff[i]/rho[i]/(epsilon[i] + SMALL), 0));
if (tk > SMALL)
{
kappa_[i] = tc[i]/(tc[i] + tk);
}
else
{
kappa_[i] = 1.0;
}
}
}
}
template<class ReactionThermo>
Foam::tmp<Foam::fvScalarMatrix>
Foam::combustionModels::PaSR<ReactionThermo>::R(volScalarField& Y) const
{
return kappa_*laminar<ReactionThermo>::R(Y);
}
template<class ReactionThermo>
Foam::tmp<Foam::volScalarField>
Foam::combustionModels::PaSR<ReactionThermo>::Qdot() const
{
return tmp<volScalarField>
(
new volScalarField
(
this->thermo().phasePropertyName(typeName + ":Qdot"),
kappa_*laminar<ReactionThermo>::Qdot()
)
);
}
template<class ReactionThermo>
bool Foam::combustionModels::PaSR<ReactionThermo>::read()
{
if (laminar<ReactionThermo>::read())
{
this->coeffs().readEntry("Cmix", Cmix_);
return true;
}
else
{
return false;
}
}
// ************************************************************************* //
Jacobian Member Function Definition in the StandardChemistryModel.C File: Code:
template<class ReactionThermo, class ThermoType>
void Foam::StandardChemistryModel<ReactionThermo, ThermoType>::jacobian
(
const scalar t,
const scalarField& c,
scalarField& dcdt,
scalarSquareMatrix& dfdc
) const
{
const scalar T = c[nSpecie_];
const scalar p = c[nSpecie_ + 1];
forAll(c_, i)
{
c_[i] = max(c[i], 0.0);
}
dfdc = Zero;
// Length of the first argument must be nSpecie_
omega(c_, T, p, dcdt);
forAll(reactions_, ri)
{
const Reaction<ThermoType>& R = reactions_[ri];
const scalar kf0 = R.kf(p, T, c_);
const scalar kr0 = R.kr(kf0, p, T, c_);
forAll(R.lhs(), j)
{
const label sj = R.lhs()[j].index;
scalar kf = kf0;
forAll(R.lhs(), i)
{
const label si = R.lhs()[i].index;
const scalar el = R.lhs()[i].exponent;
if (i == j)
{
if (el < 1.0)
{
if (c_[si] > SMALL)
{
kf *= el*pow(c_[si], el - 1.0);
}
else
{
kf = 0.0;
}
}
else
{
kf *= el*pow(c_[si], el - 1.0);
}
}
else
{
kf *= pow(c_[si], el);
}
}
forAll(R.lhs(), i)
{
const label si = R.lhs()[i].index;
const scalar sl = R.lhs()[i].stoichCoeff;
dfdc(si, sj) -= sl*kf;
}
forAll(R.rhs(), i)
{
const label si = R.rhs()[i].index;
const scalar sr = R.rhs()[i].stoichCoeff;
dfdc(si, sj) += sr*kf;
}
}
forAll(R.rhs(), j)
{
const label sj = R.rhs()[j].index;
scalar kr = kr0;
forAll(R.rhs(), i)
{
const label si = R.rhs()[i].index;
const scalar er = R.rhs()[i].exponent;
if (i == j)
{
if (er < 1.0)
{
if (c_[si] > SMALL)
{
kr *= er*pow(c_[si], er - 1.0);
}
else
{
kr = 0.0;
}
}
else
{
kr *= er*pow(c_[si], er - 1.0);
}
}
else
{
kr *= pow(c_[si], er);
}
}
forAll(R.lhs(), i)
{
const label si = R.lhs()[i].index;
const scalar sl = R.lhs()[i].stoichCoeff;
dfdc(si, sj) += sl*kr;
}
forAll(R.rhs(), i)
{
const label si = R.rhs()[i].index;
const scalar sr = R.rhs()[i].stoichCoeff;
dfdc(si, sj) -= sr*kr;
}
}
}
// Calculate the dcdT elements numerically
const scalar delta = 1.0e-3;
omega(c_, T + delta, p, dcdt_);
for (label i=0; i<nSpecie_; i++)
{
dfdc(i, nSpecie_) = dcdt_[i];
}
omega(c_, T - delta, p, dcdt_);
for (label i=0; i<nSpecie_; i++)
{
dfdc(i, nSpecie_) = 0.5*(dfdc(i, nSpecie_) - dcdt_[i])/delta;
}
dfdc(nSpecie_, nSpecie_) = 0;
dfdc(nSpecie_ + 1, nSpecie_) = 0;
}
-------------------- Thanks, GeekCFD |
|
|
|

|
|
|
|
|
#2 |
|
New Member
Harsh Anand
Join Date: May 2024
Posts: 12
Rep Power: 2  |
Any help
 on this topic would be appreciated!! on this topic would be appreciated!!-------------------- Thanks, GeekCFD |
|
|
|

|
|
|
|
|
#3 | |
|
New Member
Harsh Anand
Join Date: May 2024
Posts: 12
Rep Power: 2  |
Hey Foamers,
I have successfully solved the issue. In short, I have copied the algorithm used inside the 'jacobian()' function in StandardChemistryModel.C File; and have created the 'dfdc' ScalarSquareMatrix to store the values inside the Jacobian Matrix. The code files can be found at Drive Link for your reference (Renamed PaSR to jacobianEigenvaluesPaSR for clarity). P.S.: Quote:
|
||
|
|

|
||
 |
| Tags |
| chemical time scale, eigenvalues, jacobian matrix |
|
|
 Similar Threads
Similar Threads
|
||||
| Thread | Thread Starter | Forum | Replies | Last Post |
| How to calculate and assemble a Jacobian matrix for Compressible Euler solver? | aerosayan | Main CFD Forum | 17 | January 9, 2024 05:45 |
| [solidMechanics] Support thread for "Solid Mechanics Solvers added to OpenFOAM Extend" | bigphil | OpenFOAM CC Toolkits for Fluid-Structure Interaction | 686 | December 22, 2022 10:10 |
| [Other] Contribution a new utility: refine wall layer mesh based on yPlus field | lakeat | OpenFOAM Community Contributions | 58 | December 23, 2021 03:36 |
| Setting up Lid driven Cavity Benchmark with 1M cells for multiple cores | puneet336 | OpenFOAM Running, Solving & CFD | 11 | April 7, 2019 01:58 |
| Star cd es-ice solver error | ernarasimman | STAR-CD | 2 | September 12, 2014 01:01 |